Iron Uptake as Target for New Antimicrobials
The dearth of new antimicrobials coming to market, combined with the ever increasing concerns over antimicrobial resistance means that new avenues for drug discovery are required. Emergen Bio are developing proprietary screening systems for use in discovery of novel antimicrobials directed at iron uptake in micro-organisms.
The concept of iron deprivation as a way of preventing infection is not new, but has only recently been explored as a possible approach to help address the growing concern of antibiotic resistance. Interfering with the iron metabolic pathway, either by stopping the generation or secretion of the siderophore, or by inhibiting its subsequent uptake back into the cell, offers new target for antimicrobial discovery.
Emergen Bio offer two screening assays
Antibiotic Screening
The concept of focusing on the iron uptake pathway as an alternative target for therapeutic intervention is growing. In order to assess the effect of potential antimicrobials, it is clear that they should be investigated in both an iron-rich environment, as currently done for most antibiotics, where no siderophore generation occurs, as well as an in an iron free environment, where siderophore generation will occur1. The assay system can be used to assess the potential of a compound to inhibit iron uptake. Microorganisms are grown under iron limiting conditions for 24 hours in presence of potential inhibitors. Following addition of proprietary reagents, results are available in less than 5 minutes.
Antimicrobial Discovery-Siderophore Inhibition Assay
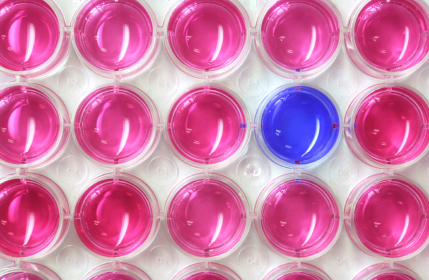
The assay is based on a reaction between a dye reagent which is blue and the siderophore. In the presence of siderophore, the dye changes colour from blue to pink, depending on the concentration of the siderophore. In the presence of an inhibitor of siderophore generation, no colour change will occur.
1: most if not all Minimum Inhibitory Concentration (MIC) studies are performed on standard iron-rich media, despite the fact that the antibiotic may exert its effect in an iron-free environment. The assay may also have application in assessing the MIC under conditions approximating those found in vivo.
Phosphopantetheinyl transferases (PPTase) Assay
The PPTase assay provides a test to screen for inhibitors of a key enzyme involved in regulating iron uptake as well as contributing to other key metabolic pathways.
PPTase inhibition: Application of a high-throughput system for detection of potential inhibitors of specific enzymes (PPTases) involved in iron uptake process.
Enzymes involved in the iron uptake pathway are potential targets for any new antimicrobial. Many fungi and bacteria have specific enzymes, termed non-ribosomal peptide synthetases (NRPS) (and polyketide synthetase), some of which are involved in the iron uptake pathway by mediating siderophore biosynthesis. However, these enzymes must first be modified (i.e. activated) by another enzyme (phosphopantetheinyl transferase, referred to as PPTase`s) before they can synthesise siderophore. PPTase`s can activate more than one NRPS enzyme so not only do they indirectly play a role in iron uptake, but also several other metabolic pathways involved in the synthesis of microbial virulence factors (e.g. mycotoxins or immunosuppressants). The key feature regarding novel antibiotic applications are
Knock out the PPTase, shut down iron uptake, as well as other metabolic pathways.
This principal has been recognised be several research groups and is currently being exploited for use in treating TB.
We have developed a simple system, in 96 well format, which can be used to assess PPTase activity, and the effectiveness of PPTase inhibitors, using fluorescent or biotin-labelled substrates. The total assay can be run in less than 1 hour (other test format or probes can be used). PPTase inhibitors will reduce or eliminate signal.
PPTase Assay Principle
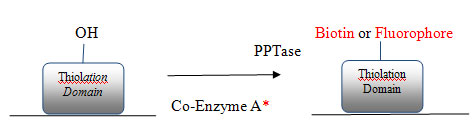
Figure 2: In the present technology, a recombinant 4'-phosphopantentheinyl transferase (PPTase) is used to catalyse the transfer of the label (e.g., biotin or fluorophore) from labelled coenzyme A to an immobilised fungal 'thiolation' domain. * Co-Enzyme was labelled either with biotin or Fluorophore.